Help improve this workflow!
This workflow has been published but could be further improved with some additional meta data:- Keyword(s) in categories input, output, operation, topic
You can help improve this workflow by suggesting the addition or removal of keywords, suggest changes and report issues, or request to become a maintainer of the Workflow .
Snakemake workflow: 16S rRNA-seq for gastric microbiota
This is a Snakemake workflow encapusulating Qiime2 modules.
The codes are distributed in the respective folders under
workflow/
, i.e.
scripts
,
rules
, and
envs
. Define the entry point of the workflow in the
Snakefile
and the main configuration in the
./config/16S-rRNA-seq.yaml
file. The .rmd script is used to analyze and visulize Qiime2 outputs.
Project
rRNA gene sequencing of the gastric microbiota of mice
The goal of the rRNA gene sequencing was two-fold: 1) to assess potential effects of the gastrin transgene on the gastric microbiota in INS-GAS mice and 2) to address the potential effects of iron deficiency on the gastric microbiota in INS-GAS mice. Collectively, the rationale of this work was to assess the contribution of the gastric microbiota in the development of precancerous lesions that occur in INS-GAS mice due to the presence of the gastrin transgene as well as iron deficiency.
Authors
- chaodi ([email protected])
Usage
Running on new respublica by: snakemake --latency-wait 10 -j 10 -p -c "sbatch --job-name={params.jobName} --mem={params.mem} -c {threads} --time=360 -e sbatch/{params.jobName}.e -o sbatch/{params.jobName}.o"
Workflow
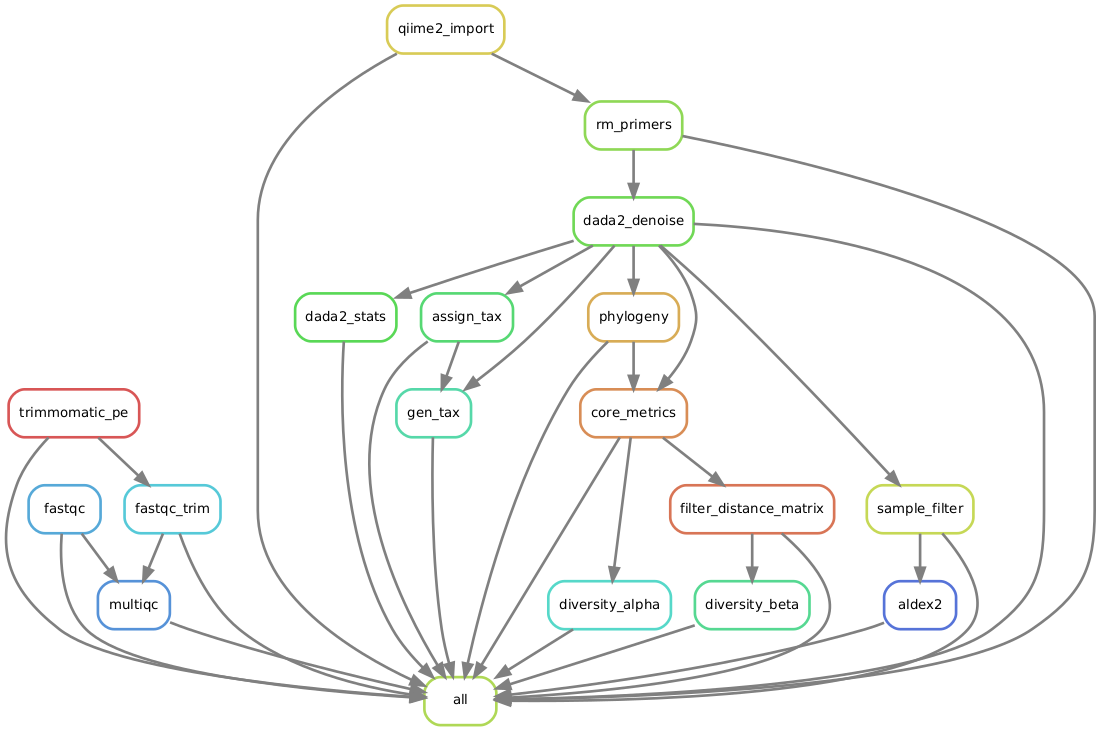
Code Snippets
71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 179 180 181 182 183 184 185 186 187 188 189 190 191 192 193 194 195 196 197 198 199 200 201 202 203 204 205 206 207 208 209 210 211 212 213 214 215 216 217 218 219 220 221 222 223 224 225 226 227 228 229 230 231 232 233 234 235 236 237 238 239 240 241 242 243 244 245 246 247 248 249 250 251 252 253 254 255 256 257 258 259 260 261 262 263 264 265 266 267 268 | shell: ''' conda activate qiime2-2019.7 #1 qiime aldex2 aldex2 \ --i-table {input.table_differentials_transgene_gastric_PicoLab_Rodent_Diet} \ --m-metadata-file {input.metadata_differentials_transgene_gastric_PicoLab_Rodent_Diet} \ --m-metadata-column Mouse_background \ --o-differentials {output.differentials_transgene_gastric_PicoLab_Rodent_Diet} \ --verbose &> {log} qiime aldex2 effect-plot \ --i-table {output.differentials_transgene_gastric_PicoLab_Rodent_Diet} \ --o-visualization {output.differentials_transgene_gastric_PicoLab_Rodent_Diet_viz} &>> {log} qiime aldex2 extract-differences \ --i-table {output.differentials_transgene_gastric_PicoLab_Rodent_Diet} \ --o-differentials {output.differentials_transgene_gastric_PicoLab_Rodent_Diet_sig} \ --p-sig-threshold 0.1 \ --p-effect-threshold 0 \ --p-difference-threshold 0 &>> {log} #2 qiime aldex2 aldex2 \ --i-table {input.table_differentials_transgene_fecal_PicoLab_Rodent_Diet} \ --m-metadata-file {input.metadata_differentials_transgene_fecal_PicoLab_Rodent_Diet} \ --m-metadata-column Mouse_background \ --o-differentials {output.differentials_transgene_fecal_PicoLab_Rodent_Diet} \ --verbose &>> {log} qiime aldex2 effect-plot \ --i-table {output.differentials_transgene_fecal_PicoLab_Rodent_Diet} \ --o-visualization {output.differentials_transgene_fecal_PicoLab_Rodent_Diet_viz} &>> {log} qiime aldex2 extract-differences \ --i-table {output.differentials_transgene_fecal_PicoLab_Rodent_Diet} \ --o-differentials {output.differentials_transgene_fecal_PicoLab_Rodent_Diet_sig} \ --p-sig-threshold 0.1 \ --p-effect-threshold 0 \ --p-difference-threshold 0 &>> {log} #3 qiime aldex2 aldex2 \ --i-table {input.table_differentials_iron_deficiency_gastric} \ --m-metadata-file {input.metadata_differentials_iron_deficiency_gastric} \ --m-metadata-column Diet_Or_Water_treatment \ --o-differentials {output.differentials_iron_deficiency_gastric} \ --verbose &>> {log} qiime aldex2 effect-plot \ --i-table {output.differentials_iron_deficiency_gastric} \ --o-visualization {output.differentials_iron_deficiency_gastric_viz} &>> {log} qiime aldex2 extract-differences \ --i-table {output.differentials_iron_deficiency_gastric} \ --o-differentials {output.differentials_iron_deficiency_gastric_sig} \ --p-sig-threshold 0.1 \ --p-effect-threshold 0 \ --p-difference-threshold 0 &>> {log} #4 qiime aldex2 aldex2 \ --i-table {input.table_differentials_iron_deficiency_fecal} \ --m-metadata-file {input.metadata_differentials_iron_deficiency_fecal} \ --m-metadata-column Diet_Or_Water_treatment \ --o-differentials {output.differentials_iron_deficiency_fecal} \ --verbose &>> {log} qiime aldex2 effect-plot \ --i-table {output.differentials_iron_deficiency_fecal} \ --o-visualization {output.differentials_iron_deficiency_fecal_viz} &>> {log} qiime aldex2 extract-differences \ --i-table {output.differentials_iron_deficiency_fecal} \ --o-differentials {output.differentials_iron_deficiency_fecal_sig} \ --p-sig-threshold 0.1 \ --p-effect-threshold 0 \ --p-difference-threshold 0 &>> {log} #5 qiime aldex2 aldex2 \ --i-table {input.table_differentials_DCA_gastric_uninfected} \ --m-metadata-file {input.metadata_differentials_DCA_gastric_uninfected} \ --m-metadata-column Diet_Or_Water_treatment \ --o-differentials {output.differentials_DCA_gastric_uninfected} \ --verbose &>> {log} qiime aldex2 effect-plot \ --i-table {output.differentials_DCA_gastric_uninfected} \ --o-visualization {output.differentials_DCA_gastric_uninfected_viz} &>> {log} qiime aldex2 extract-differences \ --i-table {output.differentials_DCA_gastric_uninfected} \ --o-differentials {output.differentials_DCA_gastric_uninfected_sig} \ --p-sig-threshold 0.25 \ --p-effect-threshold 0 \ --p-difference-threshold 0 &>> {log} #6 qiime aldex2 aldex2 \ --i-table {input.table_differentials_DCA_fecal_uninfected} \ --m-metadata-file {input.metadata_differentials_DCA_fecal_uninfected} \ --m-metadata-column Diet_Or_Water_treatment \ --o-differentials {output.differentials_DCA_fecal_uninfected} \ --verbose &>> {log} qiime aldex2 effect-plot \ --i-table {output.differentials_DCA_fecal_uninfected} \ --o-visualization {output.differentials_DCA_fecal_uninfected_viz} &>> {log} qiime aldex2 extract-differences \ --i-table {output.differentials_DCA_fecal_uninfected} \ --o-differentials {output.differentials_DCA_fecal_uninfected_sig} \ --p-sig-threshold 0.5 \ --p-effect-threshold 0 \ --p-difference-threshold 0 &>> {log} #7 qiime aldex2 aldex2 \ --i-table {input.table_differentials_infection_gastric_H20} \ --m-metadata-file {input.metadata_differentials_infection_gastric_H20} \ --m-metadata-column Infection \ --o-differentials {output.differentials_infection_gastric_H20} \ --verbose &>> {log} qiime aldex2 effect-plot \ --i-table {output.differentials_infection_gastric_H20} \ --o-visualization {output.differentials_infection_gastric_H20_viz} &>> {log} qiime aldex2 extract-differences \ --i-table {output.differentials_infection_gastric_H20} \ --o-differentials {output.differentials_infection_gastric_H20_sig} \ --p-sig-threshold 1 \ --p-effect-threshold 0 \ --p-difference-threshold 0 &>> {log} #8 qiime aldex2 aldex2 \ --i-table {input.table_differentials_infection_gastric_DCA} \ --m-metadata-file {input.metadata_differentials_infection_gastric_DCA} \ --m-metadata-column Infection \ --o-differentials {output.differentials_infection_gastric_DCA} \ --verbose &>> {log} qiime aldex2 effect-plot \ --i-table {output.differentials_infection_gastric_DCA} \ --o-visualization {output.differentials_infection_gastric_DCA_viz} &>> {log} qiime aldex2 extract-differences \ --i-table {output.differentials_infection_gastric_DCA} \ --o-differentials {output.differentials_infection_gastric_DCA_sig} \ --p-sig-threshold 0.25 \ --p-effect-threshold 0 \ --p-difference-threshold 0 &>> {log} #9 qiime aldex2 aldex2 \ --i-table {input.table_differentials_infection_fecal_H20} \ --m-metadata-file {input.metadata_differentials_infection_fecal_H20} \ --m-metadata-column Infection \ --o-differentials {output.differentials_infection_fecal_H20} \ --verbose &>> {log} qiime aldex2 effect-plot \ --i-table {output.differentials_infection_fecal_H20} \ --o-visualization {output.differentials_infection_fecal_H20_viz} &>> {log} qiime aldex2 extract-differences \ --i-table {output.differentials_infection_fecal_H20} \ --o-differentials {output.differentials_infection_fecal_H20_sig} \ --p-sig-threshold 0.25 \ --p-effect-threshold 0 \ --p-difference-threshold 0 &>> {log} #10 qiime aldex2 aldex2 \ --i-table {input.table_differentials_infection_fecal_DCA} \ --m-metadata-file {input.metadata_differentials_infection_fecal_DCA} \ --m-metadata-column Infection \ --o-differentials {output.differentials_infection_fecal_DCA} \ --verbose &>> {log} qiime aldex2 effect-plot \ --i-table {output.differentials_infection_fecal_DCA} \ --o-visualization {output.differentials_infection_fecal_DCA_viz} &>> {log} qiime aldex2 extract-differences \ --i-table {output.differentials_infection_fecal_DCA} \ --o-differentials {output.differentials_infection_fecal_DCA_sig} \ --p-sig-threshold 0.25 \ --p-effect-threshold 0 \ --p-difference-threshold 0 &>> {log} #11 qiime aldex2 aldex2 \ --i-table {input.table_differentials_4groups_gastric} \ --m-metadata-file {input.metadata_differentials_4groups_gastric} \ --m-metadata-column Diet_Or_Water_treatment \ --o-differentials {output.differentials_4groups_gastric} \ --verbose &>> {log} qiime aldex2 effect-plot \ --i-table {output.differentials_4groups_gastric} \ --o-visualization {output.differentials_4groups_gastric_viz} &>> {log} qiime aldex2 extract-differences \ --i-table {output.differentials_4groups_gastric} \ --o-differentials {output.differentials_4groups_gastric_sig} \ --p-sig-threshold 1 \ --p-effect-threshold 0 \ --p-difference-threshold 0 &>> {log} #12 qiime aldex2 aldex2 \ --i-table {input.table_differentials_4groups_fecal} \ --m-metadata-file {input.metadata_differentials_4groups_fecal} \ --m-metadata-column Diet_Or_Water_treatment \ --o-differentials {output.differentials_4groups_fecal} \ --verbose &>> {log} qiime aldex2 effect-plot \ --i-table {output.differentials_4groups_fecal} \ --o-visualization {output.differentials_4groups_fecal_viz} &>> {log} qiime aldex2 extract-differences \ --i-table {output.differentials_4groups_fecal} \ --o-differentials {output.differentials_4groups_fecal_sig} \ --p-sig-threshold 0.25 \ --p-effect-threshold 0 \ --p-difference-threshold 0 &>> {log} conda deactivate ''' |
18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 | shell: ''' conda activate qiime2-2019.7 # collapse to asv to taxon qiime taxa collapse \ --i-table {input.table_2k} \ --i-taxonomy {input.taxonomy} \ --o-collapsed-table {output.uniform_table} \ --p-level 7 &> {log} # means that we group at species level # filter feature table qiime feature-table filter-features \ --i-table {output.uniform_table} \ --p-min-frequency {config[min-frequency_all]} \ --p-min-samples {config[min-samples]} \ --o-filtered-table {output.filtered_uniform_table} &>> {log} # add a pseudocount to the FeatureTable[Frequency] qiime composition add-pseudocount \ --i-table {output.filtered_uniform_table} \ --o-composition-table {output.cfu_table} &> {log} qiime composition ancom \ --i-table {output.cfu_table} \ --m-metadata-file {input.metadata} \ --m-metadata-column study_group \ --o-visualization {output.ancom_uniform_study_group} &>> {log} qiime composition ancom \ --i-table {output.cfu_table} \ --m-metadata-file {input.metadata} \ --m-metadata-column study_day \ --o-visualization {output.ancom_uniform_study_day} &>> {log} conda deactivate ''' |
14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 | shell: ''' conda activate qiime2-2019.7 qiime dada2 denoise-paired \ --i-demultiplexed-seqs {input.q2_primerRM} \ --p-trunc-len-f {config[truncation_len-f]} \ --p-trunc-len-r {config[truncation_len-r]} \ --p-trim-left-f 0 \ --p-trim-left-r 0 \ --p-n-reads-learn {config[training]} \ --p-n-threads {threads} \ --p-chimera-method {config[chimera]} \ --o-table {output.table} \ --o-representative-sequences {output.seq} \ --o-denoising-stats {output.stats} --verbose &> {log} conda deactivate ''' |
47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 | shell: ''' conda activate qiime2-2019.7 qiime metadata tabulate \ --m-input-file {input.stats} \ --o-visualization {output.stats_viz} --verbose &> {log} qiime feature-table summarize \ --i-table {input.table} \ --o-visualization {output.table_viz} \ --m-sample-metadata-file {input.stats} --verbose &>> {log} qiime feature-table tabulate-seqs \ --i-data {input.seq} \ --o-visualization {output.seq_viz} --verbose &>> {log} qiime tools export \ --input-path {input.table} \ --output-path {params.outdir}/asv-table &>> {log} qiime tools export \ --input-path {input.stats} \ --output-path {params.outdir}/stats-dada2 &>> {log} conda deactivate ''' |
34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 | shell: ''' ## --p-sampling-depth should be carefully chosen by reviewing the table_summary file -asv-table.qzv conda activate qiime2-2019.7 # echo "this tis to test conda..." &> {log} qiime diversity core-metrics-phylogenetic \ --i-phylogeny {input.rooted_tree} \ --i-table {input.feature_table} \ --p-sampling-depth {config[sampling_depth]} \ --p-n-jobs {threads} \ --m-metadata-file {input.metadata} \ --o-rarefied-table {output.rarefied_table} \ --o-faith-pd-vector {output.faith_pd_vector} \ --o-observed-otus-vector {output.observed_otus_vector} \ --o-shannon-vector {output.shannon_vector} \ --o-evenness-vector {output.evenness_vector} \ --o-unweighted-unifrac-distance-matrix {output.unweighted_unifrac_distance_matrix} \ --o-weighted-unifrac-distance-matrix {output.weighted_unifrac_distance_matrix} \ --o-jaccard-distance-matrix {output.jaccard_distance_matrix} \ --o-bray-curtis-distance-matrix {output.bray_curtis_distance_matrix} \ --o-unweighted-unifrac-pcoa-results {output.unweighted_unifrac_pcoa_results} \ --o-weighted-unifrac-pcoa-results {output.weighted_unifrac_pcoa_results} \ --o-jaccard-pcoa-results {output.jaccard_pcoa_results} \ --o-bray-curtis-pcoa-results {output.bray_curtis_pcoa_results} \ --o-unweighted-unifrac-emperor {output.unweighted_unifrac_emperor} \ --o-weighted-unifrac-emperor {output.weighted_unifrac_emperor} \ --o-jaccard-emperor {output.jaccard_emperor} \ --o-bray-curtis-emperor {output.bray_curtis_emperor} &> {log} conda deactivate ''' |
81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 | shell: ''' conda activate qiime2-2019.7 qiime diversity alpha-group-significance \ --i-alpha-diversity {input.faith_pd_vector}\ --m-metadata-file {input.metadata} \ --o-visualization {output.faith_pd_group_significance} &> {log} qiime diversity alpha-group-significance \ --i-alpha-diversity {input.evenness_vector} \ --m-metadata-file {input.metadata} \ --o-visualization {output.evenness_group_significance} &>> {log} conda deactivate ''' |
138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 179 180 181 182 183 184 185 186 187 188 189 190 191 192 193 194 195 196 197 198 199 200 201 202 203 | shell: ''' conda activate qiime2-2019.7 qiime diversity beta-group-significance \ --i-distance-matrix {input.weighted_unifrac_distance_matrix_transgene_gastric_PicoLab_Rodent_Diet} \ --m-metadata-file {input.metadata_differentials_transgene_gastric_PicoLab_Rodent_Diet} \ --m-metadata-column Mouse_background \ --o-visualization {output.weighted_unifrac_significance_transgene_gastric_PicoLab_Rodent_Diet} &>> {log} qiime diversity beta-group-significance \ --i-distance-matrix {input.weighted_unifrac_distance_matrix_transgene_fecal_PicoLab_Rodent_Diet} \ --m-metadata-file {input.metadata_differentials_transgene_fecal_PicoLab_Rodent_Diet} \ --m-metadata-column Mouse_background \ --o-visualization {output.weighted_unifrac_significance_transgene_fecal_PicoLab_Rodent_Diet} &>> {log} qiime diversity beta-group-significance \ --i-distance-matrix {input.weighted_unifrac_distance_matrix_iron_deficiency_gastric} \ --m-metadata-file {input.metadata_differentials_iron_deficiency_gastric} \ --m-metadata-column Diet_Or_Water_treatment \ --o-visualization {output.weighted_unifrac_significance_iron_deficiency_gastric} &>> {log} qiime diversity beta-group-significance \ --i-distance-matrix {input.weighted_unifrac_distance_matrix_iron_deficiency_fecal} \ --m-metadata-file {input.metadata_differentials_iron_deficiency_fecal} \ --m-metadata-column Diet_Or_Water_treatment \ --o-visualization {output.weighted_unifrac_significance_iron_deficiency_fecal} &>> {log} qiime diversity beta-group-significance \ --i-distance-matrix {input.weighted_unifrac_distance_matrix_DCA_gastric_uninfected} \ --m-metadata-file {input.metadata_differentials_DCA_gastric_uninfected} \ --m-metadata-column Diet_Or_Water_treatment \ --o-visualization {output.weighted_unifrac_significance_DCA_gastric_uninfected} &>> {log} qiime diversity beta-group-significance \ --i-distance-matrix {input.weighted_unifrac_distance_matrix_DCA_fecal_uninfected} \ --m-metadata-file {input.metadata_differentials_DCA_fecal_uninfected} \ --m-metadata-column Diet_Or_Water_treatment \ --o-visualization {output.weighted_unifrac_significance_DCA_fecal_uninfected} &>> {log} qiime diversity beta-group-significance \ --i-distance-matrix {input.weighted_unifrac_distance_matrix_infection_gastric_H20} \ --m-metadata-file {input.metadata_differentials_infection_gastric_H20} \ --m-metadata-column Infection \ --o-visualization {output.weighted_unifrac_significance_infection_gastric_H20} &>> {log} qiime diversity beta-group-significance \ --i-distance-matrix {input.weighted_unifrac_distance_matrix_infection_gastric_DCA} \ --m-metadata-file {input.metadata_differentials_infection_gastric_DCA} \ --m-metadata-column Infection \ --o-visualization {output.weighted_unifrac_significance_infection_gastric_DCA} &>> {log} qiime diversity beta-group-significance \ --i-distance-matrix {input.weighted_unifrac_distance_matrix_infection_fecal_H20} \ --m-metadata-file {input.metadata_differentials_infection_fecal_H20} \ --m-metadata-column Infection \ --o-visualization {output.weighted_unifrac_significance_infection_fecal_H20} &>> {log} qiime diversity beta-group-significance \ --i-distance-matrix {input.weighted_unifrac_distance_matrix_infection_fecal_DCA} \ --m-metadata-file {input.metadata_differentials_infection_fecal_DCA} \ --m-metadata-column Infection \ --o-visualization {output.weighted_unifrac_significance_infection_fecal_DCA} &>> {log} conda deactivate ''' |
35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 | shell: ''' conda activate qiime2-2019.7 qiime diversity filter-distance-matrix \ --i-distance-matrix {input.weighted_unifrac_distance_matrix} \ --m-metadata-file {input.metadata_differentials_transgene_gastric_PicoLab_Rodent_Diet} \ --p-where "[Sample_type]='gastric tissue' AND [Diet_Or_Water_treatment] LIKE 'PicoLab Rodent Diet%'" \ --o-filtered-distance-matrix {output.weighted_unifrac_distance_matrix_transgene_gastric_PicoLab_Rodent_Diet} &> {log} qiime diversity filter-distance-matrix \ --i-distance-matrix {input.weighted_unifrac_distance_matrix} \ --m-metadata-file {input.metadata_differentials_transgene_fecal_PicoLab_Rodent_Diet} \ --p-where "[Sample_type]='fecal pellet' AND [Diet_Or_Water_treatment] LIKE 'PicoLab Rodent Diet%'" \ --o-filtered-distance-matrix {output.weighted_unifrac_distance_matrix_transgene_fecal_PicoLab_Rodent_Diet} &>> {log} qiime diversity filter-distance-matrix \ --i-distance-matrix {input.weighted_unifrac_distance_matrix} \ --m-metadata-file {input.metadata_differentials_iron_deficiency_gastric} \ --p-where "[Sample_type]='gastric tissue' AND [Diet_Or_Water_treatment] LIKE 'TestDiet%'" \ --o-filtered-distance-matrix {output.weighted_unifrac_distance_matrix_iron_deficiency_gastric} &>> {log} qiime diversity filter-distance-matrix \ --i-distance-matrix {input.weighted_unifrac_distance_matrix} \ --m-metadata-file {input.metadata_differentials_iron_deficiency_fecal} \ --p-where "[Sample_type]='fecal pellet' AND [Diet_Or_Water_treatment] LIKE 'TestDiet%'" \ --o-filtered-distance-matrix {output.weighted_unifrac_distance_matrix_iron_deficiency_fecal} &>> {log} qiime diversity filter-distance-matrix \ --i-distance-matrix {input.weighted_unifrac_distance_matrix} \ --m-metadata-file {input.metadata_differentials_DCA_gastric_uninfected} \ --p-where "[Sample_type]='gastric tissue' AND [Diet_Or_Water_treatment] LIKE '%H20%' AND [Infection]='Uninfected'" \ --o-filtered-distance-matrix {output.weighted_unifrac_distance_matrix_DCA_gastric_uninfected} &>> {log} qiime diversity filter-distance-matrix \ --i-distance-matrix {input.weighted_unifrac_distance_matrix} \ --m-metadata-file {input.metadata_differentials_DCA_fecal_uninfected} \ --p-where "[Sample_type]='fecal pellet' AND [Diet_Or_Water_treatment] LIKE '%H20%' AND [Infection]='Uninfected'" \ --o-filtered-distance-matrix {output.weighted_unifrac_distance_matrix_DCA_fecal_uninfected} &>> {log} qiime diversity filter-distance-matrix \ --i-distance-matrix {input.weighted_unifrac_distance_matrix} \ --m-metadata-file {input.metadata_differentials_infection_gastric_H20} \ --p-where "[Sample_type]='gastric tissue' AND [Diet_Or_Water_treatment] LIKE 'H20%'" \ --o-filtered-distance-matrix {output.weighted_unifrac_distance_matrix_infection_gastric_H20} &>> {log} qiime diversity filter-distance-matrix \ --i-distance-matrix {input.weighted_unifrac_distance_matrix} \ --m-metadata-file {input.metadata_differentials_infection_gastric_DCA} \ --p-where "[Sample_type]='gastric tissue' AND [Diet_Or_Water_treatment] LIKE '%DCA%'" \ --o-filtered-distance-matrix {output.weighted_unifrac_distance_matrix_infection_gastric_DCA} &>> {log} qiime diversity filter-distance-matrix \ --i-distance-matrix {input.weighted_unifrac_distance_matrix} \ --m-metadata-file {input.metadata_differentials_infection_fecal_H20} \ --p-where "[Sample_type]='fecal pellet' AND [Diet_Or_Water_treatment] LIKE 'H20%'" \ --o-filtered-distance-matrix {output.weighted_unifrac_distance_matrix_infection_fecal_H20} &>> {log} qiime diversity filter-distance-matrix \ --i-distance-matrix {input.weighted_unifrac_distance_matrix} \ --m-metadata-file {input.metadata_differentials_infection_fecal_DCA} \ --p-where "[Sample_type]='fecal pellet' AND [Diet_Or_Water_treatment] LIKE '%DCA%'" \ --o-filtered-distance-matrix {output.weighted_unifrac_distance_matrix_infection_fecal_DCA} &>> {log} conda deactivate ''' |
36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 | shell: ''' conda activate qiime2-2019.7 qiime feature-table filter-samples \ --i-table {input.table} \ --m-metadata-file {input.metadata_differentials_transgene_gastric_PicoLab_Rodent_Diet} \ --p-where "[Sample_type]='gastric tissue' AND [Diet_Or_Water_treatment] LIKE 'PicoLab Rodent Diet%'" \ --o-filtered-table {output.table_differentials_transgene_gastric_PicoLab_Rodent_Diet} &> {log} qiime feature-table filter-samples \ --i-table {input.table} \ --m-metadata-file {input.metadata_differentials_transgene_fecal_PicoLab_Rodent_Diet} \ --p-where "[Sample_type]='fecal pellet' AND [Diet_Or_Water_treatment] LIKE 'PicoLab Rodent Diet%'" \ --o-filtered-table {output.table_differentials_transgene_fecal_PicoLab_Rodent_Diet} &>> {log} qiime feature-table filter-samples \ --i-table {input.table} \ --m-metadata-file {input.metadata_differentials_iron_deficiency_gastric} \ --p-where "[Sample_type]='gastric tissue' AND [Diet_Or_Water_treatment] LIKE 'TestDiet%'" \ --o-filtered-table {output.table_differentials_iron_deficiency_gastric} &>> {log} qiime feature-table filter-samples \ --i-table {input.table} \ --m-metadata-file {input.metadata_differentials_iron_deficiency_fecal} \ --p-where "[Sample_type]='fecal pellet' AND [Diet_Or_Water_treatment] LIKE 'TestDiet%'" \ --o-filtered-table {output.table_differentials_iron_deficiency_fecal} &>> {log} qiime feature-table filter-samples \ --i-table {input.table} \ --m-metadata-file {input.metadata_differentials_DCA_gastric_uninfected} \ --p-where "[Sample_type]='gastric tissue' AND [Diet_Or_Water_treatment] LIKE '%H20%' AND [Infection]='Uninfected'" \ --o-filtered-table {output.table_differentials_DCA_gastric_uninfected} &>> {log} qiime feature-table filter-samples \ --i-table {input.table} \ --m-metadata-file {input.metadata_differentials_DCA_fecal_uninfected} \ --p-where "[Sample_type]='fecal pellet' AND [Diet_Or_Water_treatment] LIKE '%H20%' AND [Infection]='Uninfected'" \ --o-filtered-table {output.table_differentials_DCA_fecal_uninfected} &>> {log} qiime feature-table filter-samples \ --i-table {input.table} \ --m-metadata-file {input.metadata_differentials_infection_gastric_H20} \ --p-where "[Sample_type]='gastric tissue' AND [Diet_Or_Water_treatment] LIKE 'H20%'" \ --o-filtered-table {output.table_differentials_infection_gastric_H20} &>> {log} qiime feature-table filter-samples \ --i-table {input.table} \ --m-metadata-file {input.metadata_differentials_infection_gastric_DCA} \ --p-where "[Sample_type]='gastric tissue' AND [Diet_Or_Water_treatment] LIKE '%DCA%'" \ --o-filtered-table {output.table_differentials_infection_gastric_DCA} &>> {log} qiime feature-table filter-samples \ --i-table {input.table} \ --m-metadata-file {input.metadata_differentials_infection_fecal_H20} \ --p-where "[Sample_type]='fecal pellet' AND [Diet_Or_Water_treatment] LIKE 'H20%'" \ --o-filtered-table {output.table_differentials_infection_fecal_H20} &>> {log} qiime feature-table filter-samples \ --i-table {input.table} \ --m-metadata-file {input.metadata_differentials_infection_fecal_DCA} \ --p-where "[Sample_type]='fecal pellet' AND [Diet_Or_Water_treatment] LIKE '%DCA%'" \ --o-filtered-table {output.table_differentials_infection_fecal_DCA} &>> {log} qiime feature-table filter-samples \ --i-table {input.table} \ --m-metadata-file {input.metadata_differentials_4groups_gastric} \ --p-where "[Sample_type]='gastric tissue' AND [Diet_Or_Water_treatment] LIKE '%H20%'" \ --o-filtered-table {output.table_differentials_4groups_gastric} &>> {log} qiime feature-table filter-samples \ --i-table {input.table} \ --m-metadata-file {input.metadata_differentials_4groups_fecal} \ --p-where "[Sample_type]='fecal pellet' AND [Diet_Or_Water_treatment] LIKE '%H20%'" \ --o-filtered-table {output.table_differentials_4groups_fecal} &>> {log} conda deactivate ''' |
16 17 18 19 20 21 22 23 24 25 26 27 28 29 | shell: ''' conda activate qiime2-2019.7 qiime phylogeny align-to-tree-mafft-fasttree \ --p-n-threads {threads} \ --i-sequences {input.seq} \ --o-alignment {output.alignment} \ --o-masked-alignment {output.masked_alignment} \ --o-tree {output.unrooted_tree} \ --o-rooted-tree {output.rooted_tree} \ --verbose &> {log} conda deactivate ''' |
13 14 | shell: "fastqc --quiet -t {threads} --outdir ../results/fastqc {input} &> {log}" |
33 34 35 36 37 | shell: ''' trimmomatic PE -threads {threads} -phred33 -quiet {input.r1} {input.r2} \ {output.r1} {output.r1_unpaired} {output.r2} {output.r2_unpaired} {params.trimmer} ''' |
51 52 | shell: "fastqc --quiet -t {threads} --outdir ../results/fastqc_trim {input} &> {log}" |
67 68 69 70 71 72 | shell: """ multiqc --force --quiet --filename multiqc.html --outdir ../results/raw_multi_fastqc {input.raw_qc} &> {log} #run multiqc # repeat for trimmed data multiqc --force --quiet --filename multiqc.html --outdir ../results/trim_multi_fastqc {input.trim_qc} &>> {log} #run multiqc """ |
15 16 17 18 19 20 21 22 23 24 | shell: ''' # Imports demultiplexed paired end FASTQ files # Needed to create a unique manifest file to map file paths to sample ids qiime tools import \ --type 'SampleData[PairedEndSequencesWithQuality]' \ --input-path {input} \ --input-format PairedEndFastqManifestPhred33V2 \ --output-path {output.q2_import} &> {log} ''' |
14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 | shell: ''' qiime cutadapt trim-paired \ --p-cores {threads} \ --i-demultiplexed-sequences {input.q2_import} \ --p-front-f {config[primerF]} \ --p-front-r {config[primerR]} \ --p-error-rate {config[primer_err]} \ --p-overlap {config[primer_overlap]} \ --p-match-read-wildcards \ --p-match-adapter-wildcards \ --o-trimmed-sequences {output.q2_primerRM} &> {log} qiime demux summarize \ --i-data {input.q2_import} \ --o-visualization {output.q2_raw_viz} &>> {log} qiime demux summarize \ --i-data {output.q2_primerRM} \ --o-visualization {output.q2_primerRM_viz} &>> {log} ''' |
19 20 21 22 23 24 25 26 27 | shell: ''' conda activate qiime2-2019.7 qiime feature-classifier classify-sklearn \ --i-classifier {input.classifier} \ --i-reads {input.seq} \ --o-classification {output.taxonomy} &> {log} conda deactivate ''' |
43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 | shell: ''' qiime metadata tabulate \ --m-input-file {input.taxonomy} \ --o-visualization {output.taxonomy_viz} &> {log} qiime tools export \ --input-path {input.taxonomy} \ --output-path {params.outdir} &>> {log} qiime taxa barplot \ --i-table {input.feature_table} \ --i-taxonomy {input.taxonomy} \ --m-metadata-file {input.metadata} \ --o-visualization {output.taxa_barplot_viz} &>> {log} ''' |
Support
- Future updates
Related Workflows










