Help improve this workflow!
This workflow has been published but could be further improved with some additional meta data:- Keyword(s) in categories input, output, operation, topic
You can help improve this workflow by suggesting the addition or removal of keywords, suggest changes and report issues, or request to become a maintainer of the Workflow .
Table of Contents
Description
Installation
Usage
Description
This repository contains a demonstration of a basic bulk RNA-seq pipeline in two forms,
Snakemake
(in the
workflow/
directory) and as a
bash
script (in the
bash_workflow/
directory).
The two workflows perform the same analysis, but the
Snakemake
pipeline is much more robust and recommended, though not perfect just yet. This
Snakemake
pipeline was created as part of a tutorial, and thus refrains from employing some of the full/more complex utilities of
Snakemake
. Still, it is a maked improvement over the
bash
workflow.
A couple of advantages of the
Snakemake
version.
-
Stops at an error, removes incomplete files automatically
-
Can rerun/restart a pipeline after an error and only runs steps that were not completely finished
-
Can run to a certain point within the pipeline without editing the pipeline
-
Can add new samples and rerun the pipeline so that it only runs the steps necessary for processing of the new samples and integration of those new data with other sample data, if necessary
-
Can generalize the analysis
-
Do not have to pre-install all software
-
Can have temporary files removed automatically
-
With the use of a
config.yamlfile, need only to edit a single file to run a new analysis -
And more...
Steps performed
| Step | Software/Tool |
|---|---|
| Initial read QC |
FastQC
|
| Filter, trim reads |
TrimGalore!
|
| Align reads |
STAR
|
| Abundance quantification |
featureCounts
from
Subread
|
| Pair-wise differential expression analysis |
DESeq2
|
The
Snakemake
workflow structure
$ tree workflow
workflow
├── Snakefile
├── envs
│ ├── fastqc_env.yml
│ ├── star_env.yml
│ ├── subread_env.yml
│ └── trim_galore_env.yml
├── resources
│ ├── config.yaml
│ └── metadata.txt
└── scripts
└── DESeq2.R
A brief description of the files within the workflow
| File Name | Description |
|---|---|
Snakefile
|
Main file of the workflow. Import modules, creates variables, describes commands in the form of
rules
.
|
*_env.yml
files
|
.yaml
file that describes all software to be used by
conda
/
mamba
to create a computational environment, including the versions and all of the dependencies.
|
config.yaml
|
Configuration
.yaml
file holding values that we want to pass to the pipeline for anything we might want to change between runs of the pipeline, such as input file directories, parameter values, choices in trimming tools, etc.
|
metadata.txt
|
Tab-separated file containing
sample_id
and
expt
columns to be filled with the sample ids/names and their experimental group, respectively. The column names should remain unchanged. This file is used by
DESeq2
to perform a pair-wise differential expression analysis (single factor, two levels).
|
DESeq2.R
|
R script that performs the differntial expression analysis. |
config.yaml
File Field Descriptions and Example
| Field | Description | Format |
|---|---|---|
out_dir:
|
Directory to output all results. Does not need to exist yet. | Full path to directory |
(input_dir)
fastq:
|
Directory containing input reads | Full path to directory |
(input_dir)
genome_fasta:
|
Directory containing
STAR
index files
|
Full path to directory |
(input_dir)
genome_gtf:
|
GTF file used to build the
STAR
index
|
Full path to file |
(read_info)
fastq_suffix:
|
Whether the file uses
fastq
or
fq
in its name
|
String, either
fastq
or
fq
|
(read_info)
r1_suffix:
|
The string that designates read 1 files, either
_1
or
_R1
or something similar. Without any leading or trailing characters.
|
String,
_1
or
_R1
usually, no extra
.
or other characters
|
(thread_info)
fastqc:
|
Number of threads to use for each
FastQC
job
|
Integer |
(thread_info)
align:
|
Number of threads to use for each
STAR
job
|
Integer |
(thread_info)
counts:
|
Number of threads to use for each
featureCounts
job
|
Integer |
(trim_info)
length:
|
Shortest length a read can be after trimming, will toss read otherwise | Integer |
(trim_info)
quality:
|
Trim bases where quality is below this value | Integer |
(count_info)
strand:
|
Strandedness of library preparation (0=unstranded, 1=forward, 2=reverse) | Integer |
(diffex_info)
prefix:
|
String to use as a prefix for output in differential expression analysis file | Quoted string |
(diffex_info)
metadata:
|
File containing samples and group assignment for differential expression analysis |
Full path to tab-separated
.txt
file
|
Example
config.yaml
out_dir: '/Users/csifuentes/Documents/outData'
input_dir: fastq: '/Users/csifuentes/Documents/testData/simulated_reads/fastq' genome_fasta: '/Users/csifuentes/Documents/testData/index/' genome_gtf: '/Users/csifuentes/Documents/testData/index/Homo_sapiens.GRCh38.103.gtf'
read_info: fastq_suffix: 'fastq' r1_suffix: '_1'
thread_info: fastqc: 4 align: 4 counts: 4
trim_info: length: 20 quality: 30
count_info: strand: 0
diffex_info: prefix: 'WT_vs_KO' metadata: '/Users/csifuentes/Documents/repo/snakemake-demo-rnaseq/workflow/resources/metadata.txt'
Key Assumptions
-
Files are gzipped, ending with
.gz. -
Paired-end reads.
-
Only 2 groups to compare (WT vs KO, or T0 vs T24, etc.)
Installation:
-
Install miniconda using the appropriate directions from here: https://docs.conda.io/projects/conda/en/latest/user-guide/install/index.html
-
After closing and re-opening the terminal, the
condabase environment should be enabled. To check this, make sure you see(base)to the left of your user name in the terminal prompt.
(base) user: $
-
Install
mambausing conda in your base environment.
conda install -n base conda-forge mamba
-
Install
Snakemakeusingconda/mamba. I install it into it's own environment, namedsnakemake_envhere.
conda activate base
mamba create -c conda-forge -c bioconda -n snakemake_env snakemake
-
Go to https://github.com/cjsifuen/Basic-Bulk-RNA-seq-Workflow. Click the green
Codebutton on the top-right side of the page, and selectDownload ZIP, shown in the purple box below.
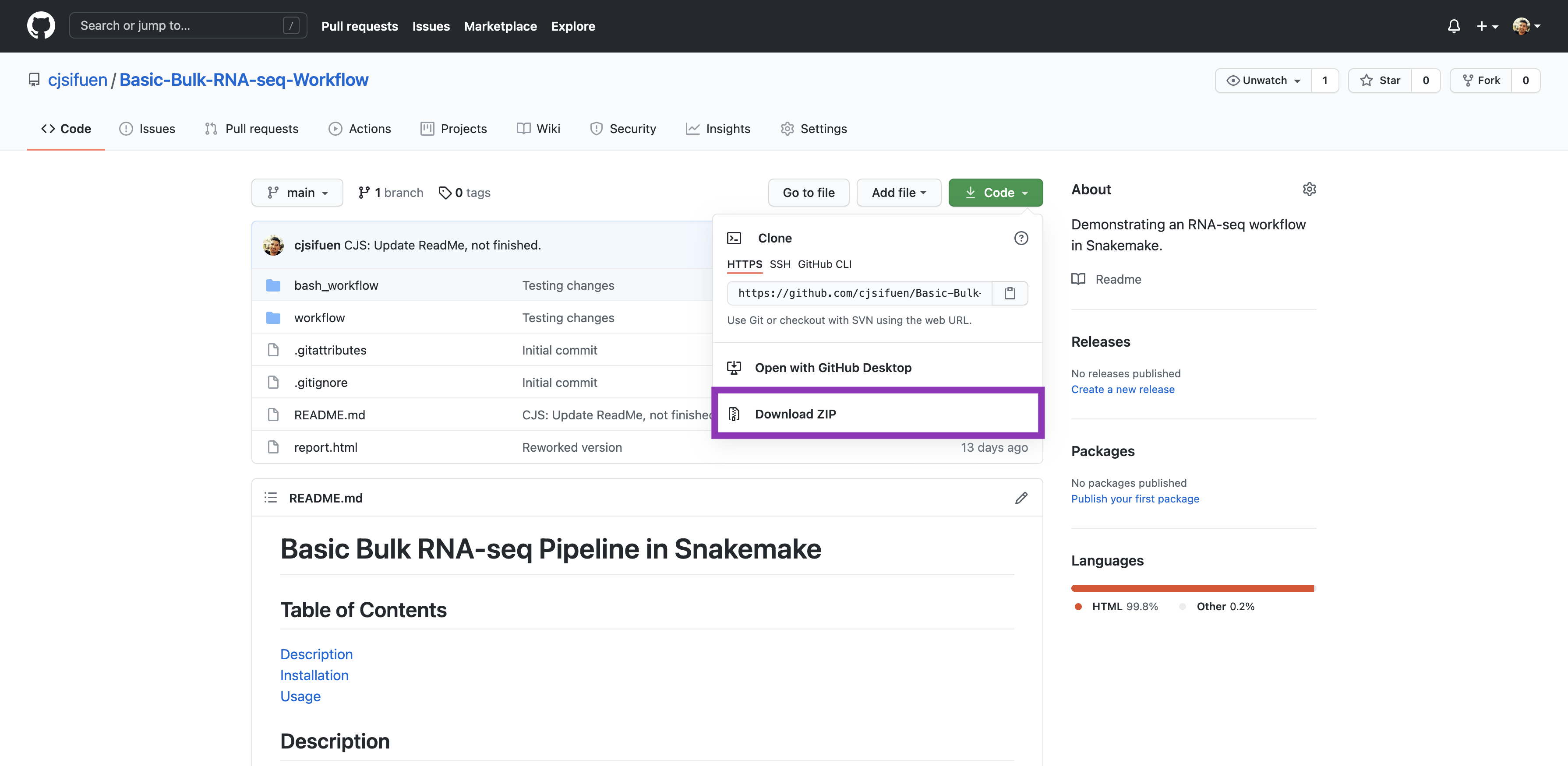
-
Move the downloaded file
Basic-Bulk-RNA-seq-Workflow-main.zipto where you want to work, and unzip (right-click and extract or unzip, or double-clicking usually works). -
If you have not installed R, install it from here: https://rstudio-education.github.io/hopr/starting.html#how-to-download-and-install-r. You can check for an R installation by opening the terminal and typing
R --version, followed by the enter key. If R is installed the version number and other information will be shown.
$ R --version
R version 4.0.3 (2020-10-10) -- "Bunny-Wunnies Freak Out"
Copyright (C) 2020 The R Foundation for Statistical Computing
Platform: x86_64-apple-darwin17.0 (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under the terms of the
GNU General Public License versions 2 or 3.
For more information about these matters see
https://www.gnu.org/licenses/.
-
Install
DESeq2in R in yoursnakemake_envby first activating thesnakemake_env. Then follow the installation instructions here: https://bioconductor.org/packages/release/bioc/html/DESeq2.html.
# activate the environment
conda activate snakemake_env
# open R
R
# install DESeq2
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("DESeq2")
-
Exit
Rby typingq(), followed by enter. Selectnfor no. Everything should be ready.
Usage:
-
Edit the
config.yamlfile, described above in description . Note the key assumptions being made about these data/files. -
Edit the
metadata.txtfile. -
Open the terminal and activate the
snakemake_env
conda activate snakemake_env
-
Change into the directory containing the unzipped repository. If I downloaded the repository to my
Downloadsdirectory and unzipped it there, I would type
cd /Users/csifuentes/Downloads/Basic-Bulk-RNA-seq-Workflow-main
- Test the pipeline to make sure everything looks okay.
snakemake -n
Should produce the following (as well as a larger block of text). This shows the number of jobs that will be run for each rule. So, for 6 samples, we have 12
fastq
files (since they are paried). Thus, I should see 12
fastqc
jobs that need to be performed. I should also see 6
star
jobs for alignment. I see these below. Everything looks good so I can continue to run the analyis.
Job counts: count jobs 1 all 1 clean_counts 1 deseq2 12 fastqc 1 featureCounts 6 star 6 trim_galore 28
This was a dry-run (flag -n). The order of jobs does not reflect the order of execution.
Check that the number of
fastqc
run
- Run the pipeline. Some useful parameters are shown below.
| Flag / option | Purpose | Required? | |
|---|---|---|---|
snakemake
|
Calls snakemake | Yes | |
--use-conda
|
Install the necessary software in temporary environments, using
conda
|
Yes | |
--conda-frontend mamba
|
Use
mamba
to install the software, which will be faster
|
No | |
--cores 8
|
Use up to 8 threads | No | |
-p
|
Print the actual command to the screen | No | |
| `-p 2>&1 | tee log.file` | Test | No |
--until star
|
Stop the pipeline after the
star
jobs are all complete
|
No |
snakemake --use-conda --conda-frontend mamba --cores 8 -p 2>&1 | tee log.file
Code Snippets
87 88 89 90 91 92 93 94 95 96 | shell: """ mkdir -p {params.qc_dir} fastqc \ --noextract \ -f {params.format} \ -o {params.qc_dir} \ -t {threads} \ {input.fastq} """ |
113 114 115 116 117 118 119 120 121 122 | shell: """ trim_galore \ -q {params.quality} \ --length {params.length} \ --basename {params.base} \ -o {params.out_dir} \ --dont_gzip \ --paired {input.r1} {input.r2} """ |
139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 | shell: """ STAR \ --genomeDir {params.index} \ --readFilesIn {input.r1} {input.r2} \ --sjdbGTFfile {params.gtf} \ --outFileNamePrefix {params.out_prefix} \ --runThreadN {threads} \ --outSAMattributes Standard \ --outSAMtype BAM SortedByCoordinate \ --outFilterMismatchNmax 999 \ --alignSJoverhangMin 8 \ --alignSJDBoverhangMin 1 \ --outFilterMismatchNoverReadLmax 0.04 \ --alignIntronMin 20 \ --alignIntronMax 1000000 \ --alignMatesGapMax 1000000 \ --sjdbScore 1 """ |
176 177 178 179 180 181 182 183 184 185 186 187 188 189 190 | shell: """ featureCounts \ -O \ -p \ -B \ -C \ -T {threads} \ -s {params.strand} \ -a {params.gtf} \ -o {output.tmp} \ {input.bams} sed '/^#/d' {output.tmp} | cut -f 1,7- > {output.counts} """ |
198 199 200 201 202 203 204 205 206 207 208 209 210 211 212 213 | run: #import modules import pandas as pd import os #read in counts file counts_df = pd.read_csv(filepath_or_buffer=input.counts, sep='\t', header=0) #grab column names, remove path and extension to get desired basenames of files fixed_names = [os.path.basename(x).split('Aligned.sortedByCoord.out.bam')[0] for x in counts_df.columns] #reassign column values of df counts_df.columns = fixed_names #write out to fixed file counts_df.to_csv(path_or_buf=output.counts, sep = '\t', index=False, header=True) |
228 229 230 231 | shell: """ Rscript {params.script} {params.metadata} {input.counts} {params.out_prefix} """ |
Support
- Future updates
Related Workflows










