COSIFER-COnSensus Interaction Network InFErence Service
COnSensus Interaction Network InFErence Service
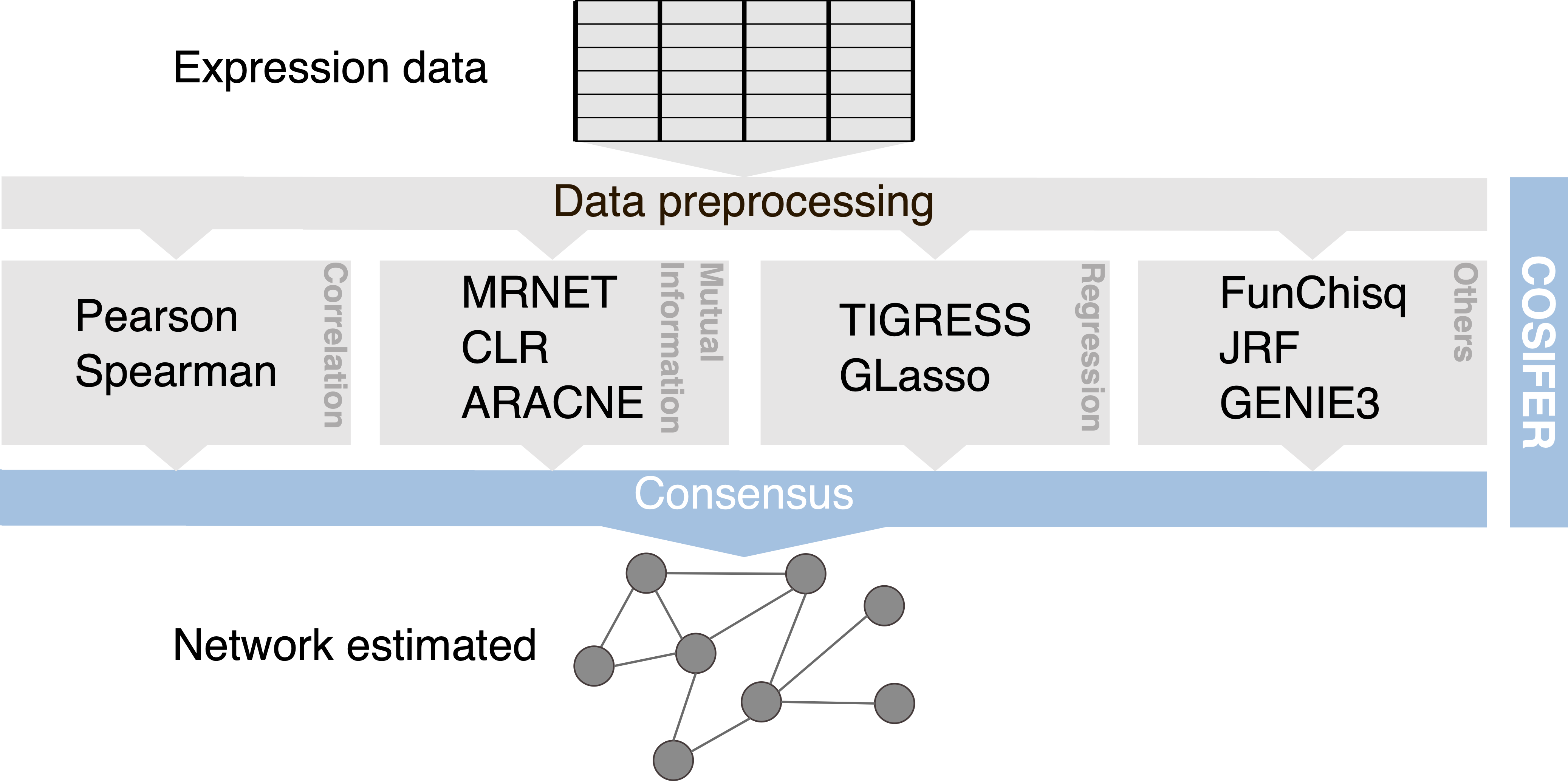
Inference framework for reconstructing networks using a consensus approach between multiple methods and data sources.
Reference
Code Snippets
34 35 36 | """ cosifer -i "${matrix}" "--sep=${separator}" ${index_col!='' ? '--index ' + index_col : ''} ${gmt_filepath.name != '.empty.' ? '--gmt_filepath ' + gmt_filepath : ''} -o "resdir" ${samples_on_rows ? '--samples_on_rows' : ''} """ |
Support
Do you know this workflow well? If so, you can
request seller status , and start supporting this workflow.
Created: 1yr ago
Updated: 1yr ago
Maitainers:
public
URL:
https://github.com/inab/ipc_workflows/tree/main/cosifer/nextflow
Name:
cosifer-1
Version:
Version 1
Downloaded:
0
Copyright:
Public Domain
License:
Boost Software License 1.0
- Future updates
Related Workflows

ENCODE pipeline for histone marks developed for the psychENCODE project
psychip pipeline is an improved version of the ENCODE pipeline for histone marks developed for the psychENCODE project.
The o...

Near-real time tracking of SARS-CoV-2 in Connecticut
Repository containing scripts to perform near-real time tracking of SARS-CoV-2 in Connecticut using genomic data. This pipeli...

snakemake workflow to run cellranger on a given bucket using gke.
A Snakemake workflow for running cellranger on a given bucket using Google Kubernetes Engine. The usage of this workflow ...

ATLAS - Three commands to start analyzing your metagenome data
Metagenome-atlas is a easy-to-use metagenomic pipeline based on snakemake. It handles all steps from QC, Assembly, Binning, t...
raw sequence reads
Genome assembly
Annotation track
checkm2
gunc
prodigal
snakemake-wrapper-utils
MEGAHIT
Atlas
BBMap
Biopython
BioRuby
Bwa-mem2
cd-hit
CheckM
DAS
Diamond
eggNOG-mapper v2
MetaBAT 2
Minimap2
MMseqs
MultiQC
Pandas
Picard
pyfastx
SAMtools
SemiBin
Snakemake
SPAdes
SqueezeMeta
TADpole
VAMB
CONCOCT
ete3
gtdbtk
h5py
networkx
numpy
plotly
psutil
utils
metagenomics

RNA-seq workflow using STAR and DESeq2
This workflow performs a differential gene expression analysis with STAR and Deseq2. The usage of this workflow is described ...

This Snakemake pipeline implements the GATK best-practices workflow
This Snakemake pipeline implements the GATK best-practices workflow for calling small germline variants. The usage of thi...





